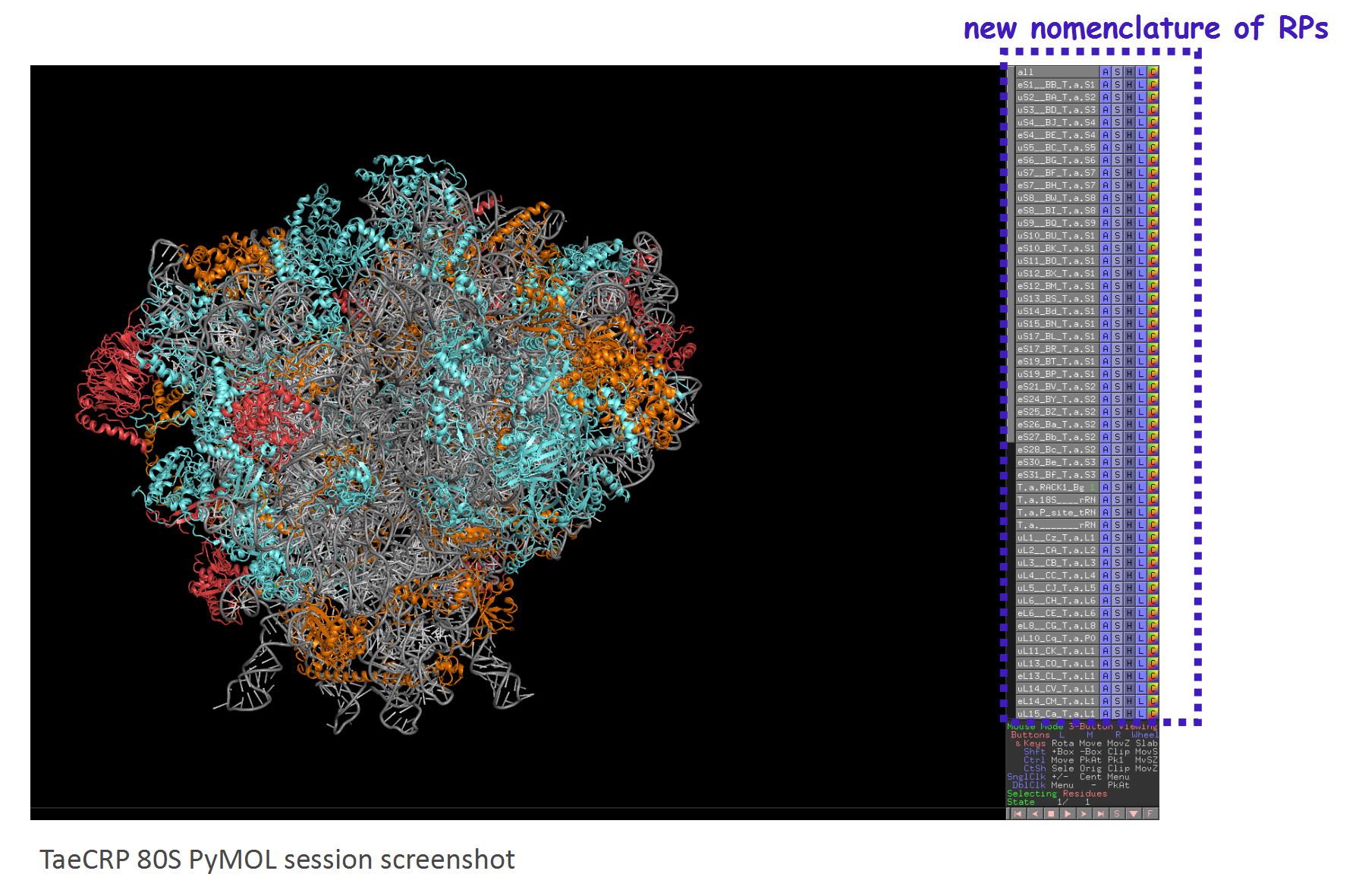
We labeled the RP subunits on the architecture of Triticum aestivum 80S ribosome according to the new nomenclature.
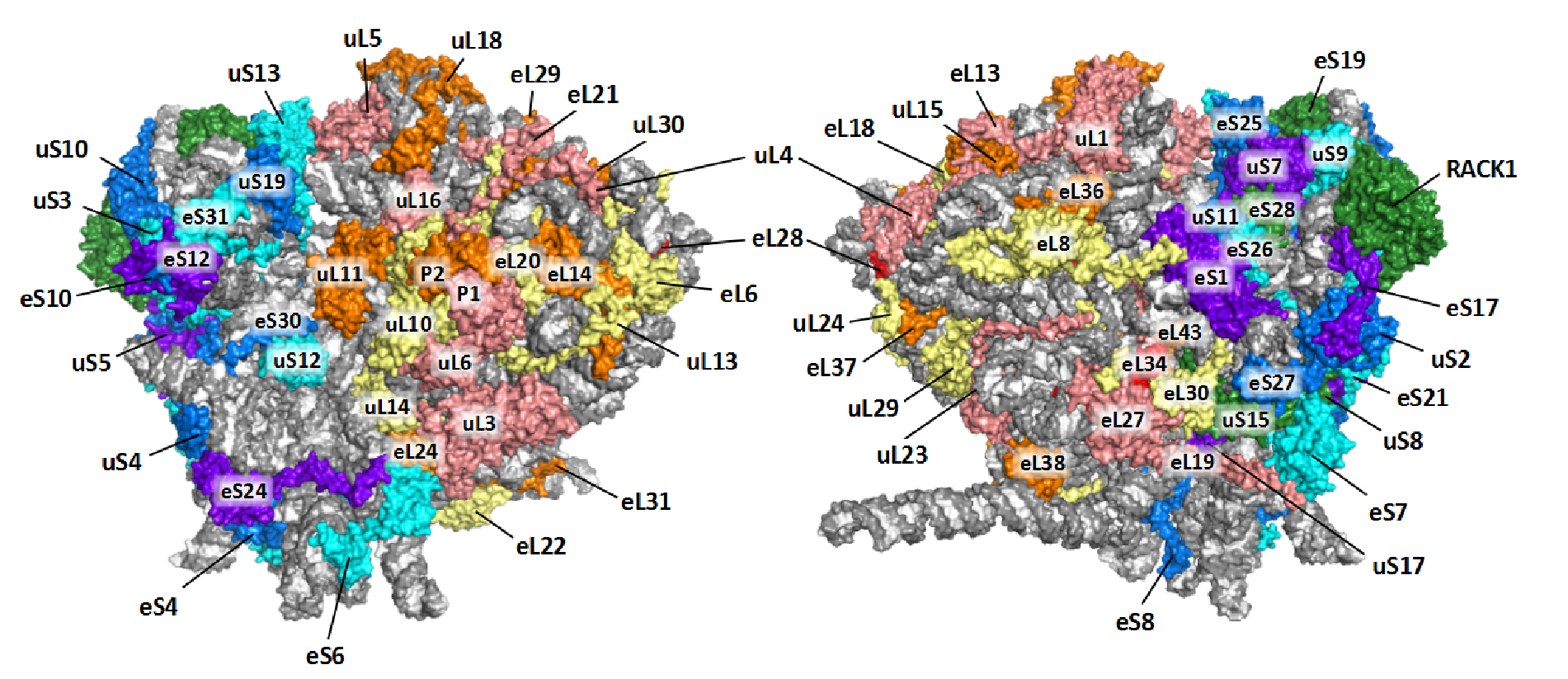
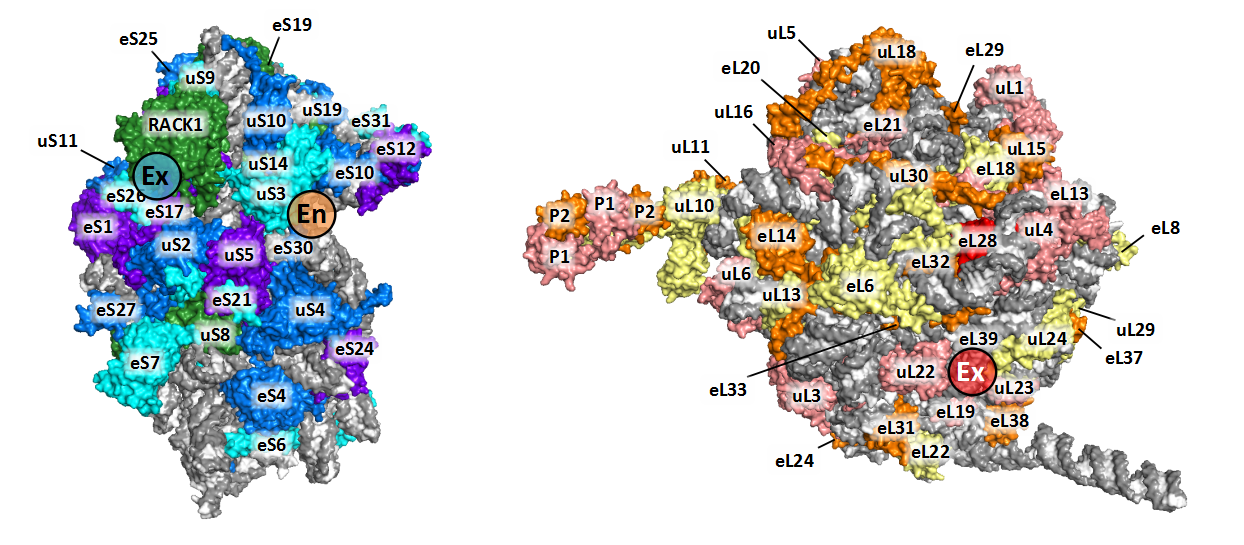
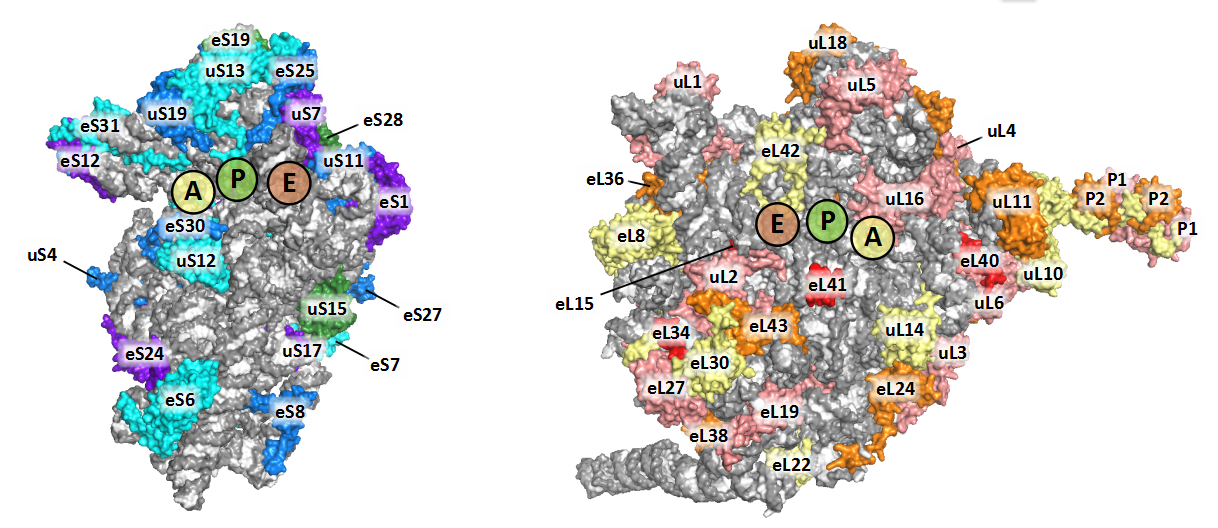
If you want display the structure yourself, you could download our TaeCRP 80S PyMOL session file and analysis it using PyMOL (version 2.2 or higher). If you want to get more annotated structures for cytoplasmic ribosomes from bacteria, yeast and human, as well as mitochondrial and chloroplast ribosomes, you could visit the Ban Lab.
Reference:
Gogala M. et al. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature. 2014. 506(7486): 107–110. DOI: https://doi.org/10.1038/nature12950
Armache, J. P. et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5 Å resolution. Proc. Natl Acad. Sci. USA. 2010. 107: 19748–19753. DOI: https://doi.org/10.1073/pnas.1009999107
TaeCRP 80S PyMOL Script was followed the scripts provided by the Ban Lab.